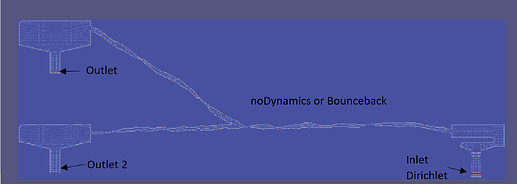
Hi Palabos Community,
I’m trying to write a code that will take my STL perform normal D3Q19 incBGKdynamics for the fluid and then pass the resulting velocity field to do Advection diffusion on some solute.
For the most part my code is working and is based on a mixture of what other people have posted here and the examples.
The major issue that I have been running into is with the Advection-Diffusion boundary conditions.
No matter how I seem to set up the BC there appears to be a layer of solute near the walls and I need to get rid of this.
I had this code working at the end of last year but something changed and I can’t for the life of me figure out what it is. Odds are i’m missing something simple and just been staring at this code too long…
Any suggestions are welcome.
The Code is below if anyone wants to attempt to read it… (it’s a mess and i’m sorry…  )
)
edit: uploaded code to a public github repo for easier reading https://github.com/Catsgomeowmeow/PalabosAD/blob/master/BiFrac.cpp
//include all palabos stuff
#include "palabos3D.h"
#include "palabos3D.hh"
//for basic math functions
#include <cmath>
using namespace plb;
//Defining the unit type
typedef double T;
typedef Array<T,3> Velocity;
// Descriptors of the LBM scheme
#define DESCRIPTOR descriptors::D3Q19Descriptor
#define CONCENTRATION_DESCRIPTOR descriptors::AdvectionDiffusionD3Q7Descriptor
// define the constant variables
plint extraLayer = 1; // Make the bounding box larger; for visualization purposes
// only. For the simulation, it is OK to have extraLayer=0.
const plint blockSize = 0; // Zero means: no sparse representation.
const plint extendedEnvelopeWidth = 0; // Because the Guo off lattice boundary condition
const plint envelopeWidth = 1;
//Booleans from the param sheet
bool useParallelIO;
bool performOutput = true; //Creates the VTK files when True
bool doImages = true;
bool saveDynamicContent; // Save dynamic content in the checkpoint files or not?
bool reverse = false ;
bool doConcentration = true;
bool doFlow = true;
bool correct = true;
bool flowContinuous =true;
bool onEuler = false;
bool flow ; // old parameter
// Other Variables from the input file
std::string meshFileName;//STL file name
TriangleSet<T>* triangleSet =0; //triangle sets for mesh
plint maxIter = 0.; // Max iteration for flow
plint maxcIter = 0.; // max iteration for solute
plint referenceResolution = 0.; //resolution of the mesh
T flowPercent = 0. ; // Flow distribution scheme
plint referenceDirection = 0.; //Direction of the model
T nuLB = 0; //kinematic viscosity in lb units
T omega = 0; //relaxation time
T concentrationOmega = 0; //relaxation time for solute
T fluidDensity = 0.; // fluid density of water (real units )
T kinematicViscosity; //kinematic viscosity in real units
T dt, dx; // Time and space steps
T D ; //diffusion coefficient
T Pe; //peclet number
T pulse_length = 31000; //pulse length for solute (should be variable )
plint interval = 0; //image interval for fluid
plint cinterval = 0; // image interval for solute
std::string name = " "; //file name
T aperture = 0.; // mean aperture in real units
void readParameters(XMLreader const& document)
{ std::string meshFileName;
document["geometry"]["mesh"].read(meshFileName); // the name of the STL FILE
document ["geometry"]["aperture"].read(aperture); // for calculation of reynolds number
document["fluid"]["difcoef"].read(D); // diffusion coffecient
document["fluid"]["kinematicViscosity"].read(kinematicViscosity);
document["fluid"]["density"].read(fluidDensity);
document["fluid"]["pe"].read(Pe);
document["fluid"]["flowPercent"].read(flowPercent);
document["numerics"]["nuLB"].read(nuLB);
document["numerics"]["referenceResolution"].read(referenceResolution);
document["simulation"]["onEuler"].read(onEuler);
document["simulation"]["doImages"].read(doImages);
document["simulation"]["doConcentration"].read(doConcentration);
document["simulation"]["doFlow"].read(doFlow);
document["simulation"]["useParallelIO"].read(useParallelIO);
document["simulation"]["maxIter"].read(maxIter);
document["simulation"]["maxcIter"].read(maxcIter);
document["simulation"]["interval"].read(interval);
document["simulation"]["cinterval"].read(cinterval);
document["simulation"]["reverse"].read(reverse);
document["simulation"]["dt"].read(dt);
triangleSet = new TriangleSet<T>(meshFileName, DBL);
pcout<<"reading STL file: "<< meshFileName<<std::endl;
}
// VTK Writing Stuff
void writeImages (
MultiBlockLattice3D<T,DESCRIPTOR>& lattice,
MultiBlockLattice3D <T,CONCENTRATION_DESCRIPTOR>& clattice,
MultiScalarField3D<int>& flagMatrix,
Box3D const& vtkDomain, std::string fname, Array<T,3> location, T dx, T dt ,bool flow)
{
VtkImageOutput3D<T> vtkOut(fname, dx, location);
if (flow == false){
vtkOut.writeData<float>(*computeDensity(clattice, vtkDomain), "Concentration", fluidDensity);
vtkOut.writeData<3,float>(*computeVelocity(clattice,vtkDomain),"velocity",dx/dt);
}else{
vtkOut.writeData<3,float>(*computeVelocity(lattice,vtkDomain),"velocity",dx/dt);
}
vtkOut.writeData<float>(*copyConvert<int,T>(*extractSubDomain(flagMatrix, vtkDomain)), "flag", 1.);
}
void writeImages (
MultiBlockLattice3D<T,DESCRIPTOR>& lattice,
MultiBlockLattice3D <T,CONCENTRATION_DESCRIPTOR>& clattice,
MultiScalarField3D<int>& flag,
plint level, Array<T,3> location, T dx, T dt ,std::string name, bool flow)
{
plint nx = lattice.getNx();
plint ny = lattice.getNy();
plint nz = lattice.getNz();
Box3D yz_vtkDomain (
0, nx-1,
0, ny-1, 0, nz-1 );
writeImages(lattice, clattice, flag, yz_vtkDomain, name+"Tubless_"+util::val2str(level), location, dx, dt,flow);
}
// Domain Setup and simulation start point
void run(std::string continueFileName)
{
plint margin =0; // extra cellls around the boundary
plint borderWidth = 0; //this is for guo boundary which needs one cell, if using this margin >= border width
//This is for mesh refinement which is nice incase the input mesh doesn't have the 4 lattice space that is required
//Reference resolution should be the input resolution
// Need to add the level part of do it manually
plint resolution = referenceResolution * util::twoToThePower(0);
/// -------------------------------------------- ///
/// --------------- Mesh reading --------------- ///
/// -------------------------------------------- ///
DEFscaledMesh<T>* defMesh =
new DEFscaledMesh<T>(*triangleSet, resolution, referenceDirection, margin, extraLayer);
TriangleBoundary3D<T> boundary(*defMesh);
delete defMesh;
boundary.getMesh().inflate();
Array<T,3> location(boundary.getPhysicalLocation());
// Calculate all the boundries in lb units
T pe = Pe;
pcout << "Peclet Number picked = " << pe << std::endl;
T vel = (pe * D)/ aperture;
T dx = boundary.getDx();
T nu = kinematicViscosity;
T nuLB = (nu*dt)/(dx*dx);
T DLB = (D*dt)/(dx*dx);
T omega = 1./(3*nuLB+0.5);
T concentrationOmega = 1./(3*DLB+0.5);
T uAveLB = vel * dt /dx;
T tau = 1./omega;
T conctau = 1./concentrationOmega;
//Debugging outputs
pcout << "Diffusion coefficient = " << D << std::endl;
pcout<< "Tau = "<<tau<< " at a dt of: "<< dt<< " and a dx of: "<< dx<<std::endl;
pcout<<"Concentration tau= "<<conctau<< std::endl;
pcout<< "Velocity from input file: " << vel<<"mm/s"<<std::endl;
pcout<<"Current average velocity in LBunits is: "<< uAveLB << std::endl;
pcout<< "Reynolds number based on an aperture of 0.8 mm : " << vel*aperture/kinematicViscosity << std::endl;
// Voxelize the domain and decide what is inside and what is outside
pcout << std::endl << "Voxelizing the simulation domain" << std::endl;
int flowType =voxelFlag::inside;
VoxelizedDomain3D<T> voxelizedDomain (
boundary, flowType, extraLayer, borderWidth, extendedEnvelopeWidth, blockSize );
MultiScalarField3D<int> flagMatrix((MultiBlock3D&)voxelizedDomain.getVoxelMatrix());
setToConstant(flagMatrix, voxelizedDomain.getVoxelMatrix(),
voxelFlag::inside, flagMatrix.getBoundingBox(), 1);
// setToConstant(flagMatrix, voxelizedDomain.getVoxelMatrix(),
// voxelFlag::innerBorder, flagMatrix.getBoundingBox(), 1);
// setToConstant(flagMatrix, voxelizedDomain.getVoxelMatrix(),
// voxelFlag::outerBorder, flagMatrix.getBoundingBox(), 1);
pcout << "Number of fluid cells: " << computeSum(flagMatrix) << std::endl;
//Outputs some infromation about the mesh
if (performOutput) {
pcout << getMultiBlockInfo(voxelizedDomain.getVoxelMatrix()) << std::endl;
}
//Real Position of the inlet and outlet
Array<T,3> inletRealPos(51.18297576904297, 10.5,5);
Array<T,3> outlet1RealPos(51.18297576904297,127.5,33); // this is the birfurcating outlet
Array<T,3> outlet2RealPos(51.18297576904297,127.5,5); // this is the continous outlet
T diameterReal = 1.2;
pcout << "Location Coordinates are: " << location[0]<<","<<location[1]<<","<<location[2] << std::endl;
pcout << "dx is: " << dx <<" dt is "<<dt<< std::endl;
Array<T,3> inletPos((inletRealPos-location)/dx);
Array<T,3> outlet1Pos((outlet1RealPos-location)/dx);
Array<T,3> outlet2Pos((outlet2RealPos-location)/dx);
plint diameter = util::roundToInt(diameterReal/dx);
Box3D inletDomain(util::roundToInt(inletPos[0]-diameter), util::roundToInt(inletPos[0]+diameter),
util::roundToInt(inletPos[1]-diameter), util::roundToInt(inletPos[1]+diameter),
util::roundToInt(inletPos[2]), util::roundToInt(inletPos[2]));
Box3D behindInlet(inletDomain.x0, inletDomain.x1,
inletDomain.y0, inletDomain.y1,
inletDomain.z0-1, inletDomain.z1-1);
///////////////////////////
//Bifurcating Fracture//
//////////////////////////
Box3D outlet1Domain(util::roundToInt(outlet1Pos[0]-diameter), util::roundToInt(outlet1Pos[0]+diameter),
util::roundToInt(outlet1Pos[1]-diameter), util::roundToInt(outlet1Pos[1]+diameter),
util::roundToInt(outlet1Pos[2]), util::roundToInt(outlet1Pos[2]));
Box3D behindOutlet1(outlet1Domain.x0-2, outlet1Domain.x1+2,
outlet1Domain.y0-6, outlet1Domain.y1+5.5,
outlet1Domain.z0-6, outlet1Domain.z1);
//////////////////////////
//Continous Fracture//
/////////////////////////
Box3D outlet2Domain(util::roundToInt(outlet2Pos[0]-diameter), util::roundToInt(outlet2Pos[0]+diameter),
util::roundToInt(outlet2Pos[1]-diameter), util::roundToInt(outlet2Pos[1]+diameter),
util::roundToInt(outlet2Pos[2]), util::roundToInt(outlet2Pos[2]));
Box3D behindOutlet2(outlet2Domain.x0-2, outlet2Domain.x1+2,
outlet2Domain.y0-6, outlet2Domain.y1+5.5,
outlet2Domain.z0-6, outlet2Domain.z1);
pcout<< "outletdomain z =" << outlet2Domain.z1 <<std::endl;
//////////////////
// Fluid lattice//
/////////////////
pcout << "Generating fluid lattice and boundary conditions." << std::endl;
Dynamics<T,DESCRIPTOR>* dynamics =0;
dynamics = new IncBGKdynamics<T,DESCRIPTOR>(omega);
// dynamics = new BGKdynamics<T,DESCRIPTOR>(omega);
MultiBlockLattice3D<T,DESCRIPTOR>* lattice = 0;
lattice = generateMultiBlockLattice<T,DESCRIPTOR>(voxelizedDomain.getVoxelMatrix(),
envelopeWidth, dynamics).release(); // release is needed here
lattice->toggleInternalStatistics(false);//for MPI
lattice->periodicity().toggleAll(false);// Periodic Boundries
/////////////////////
//BC for the fluid//
/////////////////////
OnLatticeBoundaryCondition3D<T,DESCRIPTOR>* boundaryCondition
= createLocalBoundaryCondition3D<T,DESCRIPTOR>();
// Apply the locations of the inlet and outlet and the bc conditions
if (reverse == true ){
// defineDynamics(*lattice, behindOutlet2, new NoDynamics<T,DESCRIPTOR>(0.));
// defineDynamics(*lattice, behindOutlet1, new NoDynamics<T,DESCRIPTOR>(0.));
// defineDynamics(*lattice, outlet2Domain, new IncBGKdynamics<T,DESCRIPTOR>(omega));
// defineDynamics(*lattice, outlet1Domain, new IncBGKdynamics<T,DESCRIPTOR>(omega));
// defineDynamics(*lattice, outlet2Domain, new BGKdynamics<T,DESCRIPTOR>(omega));
// defineDynamics(*lattice, outlet1Domain, new BGKdynamics<T,DESCRIPTOR>(omega));
//-------------------//
//Bifurcating outlet //
//--------------------//
boundaryCondition ->addVelocityBoundary2N(outlet1Domain ,*lattice,boundary::dirichlet);
setBoundaryVelocity(*lattice,outlet1Domain, Array<T,3>((T)0.,(T)0.,(T) -(2*uAveLB*flowPercent)));
setBoundaryDensity(*lattice,outlet1Domain,(T)0.);
pcout<<"velocity at outlet1 = "<<-(uAveLB*flowPercent) << std::endl;
//--------------------//
// Continous outlet
//--------------------//
boundaryCondition ->addVelocityBoundary2N(outlet2Domain, *lattice,boundary::dirichlet);
setBoundaryVelocity(*lattice,outlet2Domain, Array<T,3>((T)0.,(T)0.,(T) -(2*uAveLB*(1-flowPercent))));
setBoundaryDensity(*lattice,outlet2Domain,(T)0.);
pcout<<"velocity at outlet1 = "<<-(uAveLB*(1-flowPercent)) << std::endl;
//------//
// Inlet//
//-----//
boundaryCondition ->addPressureBoundary2N(inletDomain,*lattice,boundary::dirichlet);
boundaryCondition ->addVelocityBoundary2N(inletDomain, *lattice,boundary::dirichlet);
setBoundaryDensity(*lattice,inletDomain,(T)1.);
setBoundaryVelocity(*lattice,inletDomain, Array<T,3>((T)0.,(T)0.,(T) 0.04));
// defineDynamics(*lattice, behindInlet, new NoDynamics<T,DESCRIPTOR>(0.));
}else {// Forward Simulation
boundaryCondition->addVelocityBoundary2N(inletDomain, *lattice,boundary::dirichlet);
setBoundaryVelocity(*lattice, inletDomain, Array<T,3>((T)0.,(T)0,(T)uAveLB));
boundaryCondition->addPressureBoundary2N(outlet1Domain, *lattice,boundary::outflow);
setBoundaryDensity(*lattice, outlet1Domain, (T) 1.);
boundaryCondition->addPressureBoundary2N(outlet2Domain, *lattice,boundary::outflow);
setBoundaryDensity(*lattice, outlet2Domain, (T) 1.);
defineDynamics(*lattice, flagMatrix, lattice->getBoundingBox(), new BounceBack<T,DESCRIPTOR>(1.), 0);
}
// this resets all the outter area to a bounceback boundary conditions
defineDynamics(*lattice,voxelizedDomain.getVoxelMatrix(),lattice->getBoundingBox(),
new BounceBack<T,DESCRIPTOR>((T)0), voxelFlag::outside);
//////////////////////////////
// Concentration Lattice//
////////////////////////////
pcout<<"Generating Concentration lattice and boundary conditions"<< std::endl;
// MultiBlockLattice3D<T,CONCENTRATION_DESCRIPTOR>* concentrationLattice=
// new MultiBlockLattice3D<T,CONCENTRATION_DESCRIPTOR>((MultiBlock3D&) voxelizedDomain.getVoxelMatrix());
// defineDynamics(*concentrationLattice,concentrationLattice->getBoundingBox(),
// new BounceBack<T,CONCENTRATION_DESCRIPTOR>(0.));
// defineDynamics(*concentrationLattice,voxelizedDomain.getVoxelMatrix(),concentrationLattice->getBoundingBox(),
// new AdvectionDiffusionBGKdynamics<T,CONCENTRATION_DESCRIPTOR>(concentrationOmega),voxelFlag::inside);
Dynamics<T,CONCENTRATION_DESCRIPTOR>* cdynamics =0;
cdynamics = new AdvectionDiffusionBGKdynamics<T,CONCENTRATION_DESCRIPTOR>(concentrationOmega);
MultiBlockLattice3D<T,CONCENTRATION_DESCRIPTOR>* concentrationLattice = 0;
concentrationLattice = generateMultiBlockLattice<T,CONCENTRATION_DESCRIPTOR>(voxelizedDomain.getVoxelMatrix(),
envelopeWidth, cdynamics).release(); // release is needed here
concentrationLattice->toggleInternalStatistics(false);
concentrationLattice->periodicity().toggleAll(false);
//Boundry conditions for the solute
OnLatticeAdvectionDiffusionBoundaryCondition3D<T,CONCENTRATION_DESCRIPTOR>* concentrationBoundaryCondition=
createLocalAdvectionDiffusionBoundaryCondition3D <T,CONCENTRATION_DESCRIPTOR>();
// defineDynamics(*concentrationLattice,voxelizedDomain.getVoxelMatrix(),concentrationLattice->getBoundingBox(),
// new AdvectionDiffusionBGKdynamics<T,CONCENTRATION_DESCRIPTOR>(concentrationOmega),voxelFlag::innerBorder);
// defineDynamics(*concentrationLattice,voxelizedDomain.getVoxelMatrix(),concentrationLattice->getBoundingBox(),
// new NoDynamics<T,CONCENTRATION_DESCRIPTOR>);
// defineDynamics(*concentrationLattice,voxelizedDomain.getVoxelMatrix(),concentrationLattice->getBoundingBox(),
// new AdvectionDiffusionBGKdynamics<T,CONCENTRATION_DESCRIPTOR>(concentrationOmega),voxelFlag::inside);
// defineDynamics(*concentrationLattice, behindOutlet2, new NoDynamics<T,CONCENTRATION_DESCRIPTOR>(0.));
// defineDynamics(*concentrationLattice, behindOutlet1, new NoDynamics<T,CONCENTRATION_DESCRIPTOR>(0.));
// defineDynamics(*concentrationLattice, outlet2Domain, new IncBGKdynamics<T,CONCENTRATION_DESCRIPTOR>(concentrationOmega));
// defineDynamics(*concentrationLattice, outlet1Domain, new IncBGKdynamics<T,CONCENTRATION_DESCRIPTOR>(concentrationOmega));
//Inlet BC//
concentrationBoundaryCondition->addTemperatureBoundary2N(inletDomain, *concentrationLattice,boundary::density);
setBoundaryDensity(*concentrationLattice, inletDomain, (T)1);
// //Bifurcating BC//
// concentrationBoundaryCondition->addTemperatureBoundary2N(outlet1Domain, *concentrationLattice,boundary::density);
// setBoundaryDensity(*concentrationLattice, outlet1Domain, (T)0.);
// //Continous BC //
// concentrationBoundaryCondition->addTemperatureBoundary2N(outlet2Domain, *concentrationLattice,boundary::density);
// setBoundaryDensity(*concentrationLattice, outlet2Domain, (T)0.);
// this resets all the outter area to a bounceback boundary conditions
// defineDynamics(*concentrationLattice,voxelizedDomain.getVoxelMatrix(),concentrationLattice->getBoundingBox(),
// new BounceBack<T,CONCENTRATION_DESCRIPTOR>((T)0), voxelFlag::outside);
// Coupling
integrateProcessingFunctional(
new LatticeToPassiveAdvDiff3D<T,DESCRIPTOR,CONCENTRATION_DESCRIPTOR>((T) 1.),
lattice->getBoundingBox(), *lattice, *concentrationLattice, 1);
pcout<<"starting Simulation"<<std::endl;
global::timer("simulation").start();
// INITIALIZATION OF SIMULATION
std::vector<MultiBlock3D*> checkpointBlocks;
checkpointBlocks.push_back(lattice);
checkpointBlocks.push_back(concentrationLattice);
//Start from checkpoint
bool continueSimulation = false;
if (continueFileName != "") {
continueSimulation = true;
}
// Start the simulation
bool checkForErrors = true; // currently not used
bool stopProgram = false ;
plint iter = 0;
if (continueSimulation){
pcout<<"starting from checkpoint"<<std::endl;
loadState(checkpointBlocks,iter,saveDynamicContent,continueFileName);
lattice ->resetTime(iter);
concentrationLattice -> resetTime(iter);
pcout << " starting iteration : " << iter << std::endl;
concentrationLattice->initialize();
}else{
pcout<<"initializing lattices"<<std::endl;
initializeAtEquilibrium(*lattice, lattice->getBoundingBox(), (T) 1, Array<T,3>((T) 0.,(T) 0.,(T) 0.));
lattice->initialize();
initializeAtEquilibrium(*concentrationLattice, concentrationLattice->getBoundingBox(), (T) 0 , Array<T,3>((T) 0.,(T) 0.,(T) 0.));
concentrationLattice->initialize();
}
// Stream and collide
pcout<<"Streaming and Colliding"<<std::endl;
pcout<<"from iteration "<<iter<<std::endl;
flow = true;
for (plint i = iter; i < maxIter+1 && !stopProgram; ++i){
lattice->collideAndStream();
if (doFlow == false){
pcout<<"skipping Flow Simulation"<<std::endl;
break;
}
if (i % interval == 0 && doImages == true && i > 0){
writeImages(*lattice,*concentrationLattice,flagMatrix,i,location,dx,dt,"flow",flow);
pcout<<"saved VTI for dt = "<<i<<std::endl;
pcout<< "time to interation "<<i<< ": "<<global::timer("simulation").getTime()<<std::endl;
// pcout<< " Real time "<< i*dt
}
//save Checkpoint at last time step
if (i == maxIter ){
saveState(checkpointBlocks,i,true,"continueparam.xml","checkpoint",8); // <--- This saves 3 files
pcout<<"simulation finished in > "<< global::timer("simulation").getTime()<<std::endl;
pcout<< "saved checkpoint"<<std::endl;
}
if (i % 100000 == 0 && i > 1 ){
saveState(checkpointBlocks,i,true,"continueparam_"+std::to_string(i)+".xml","checkpoint",8); // <--- This saves 3 files
pcout<< "saved interim checkpoint"<<std::endl;
}
}
if (doConcentration == false){
pcout<< "Skipping concentration simulation" << std::endl;
return ;
}
//Currently Not needed
// Coupling of the concentration to the fluid velocity
// applyProcessingFunctional(
// new LatticeToPassiveAdvDiff3D<T,DESCRIPTOR,CONCENTRATION_DESCRIPTOR>(50.),
// concentrationLattice->getBoundingBox(),*lattice,*concentrationLattice);
// integrateProcessingFunctional(
// new LatticeToPassiveAdvDiff3D<T,DESCRIPTOR,CONCENTRATION_DESCRIPTOR>((T) 10.),
// lattice->getBoundingBox(), *lattice, *concentrationLattice, 1);
// start concentration loop
plint citer = 0;
flow = false;
pcout<<"Starting Concentration simulation for : " << maxcIter<< std::endl;
for (plint ci = citer; ci < maxcIter && !stopProgram; ++ci){
bool output = (ci == maxcIter -1 ) || stopProgram;
concentrationLattice-> collideAndStream();
if (ci% cinterval == 0 && doImages == true && ci > 0){
writeImages(*lattice,*concentrationLattice,flagMatrix,ci,location,dx,dt,"concentration",flow);
pcout<<"saved Concentration VTI "<<ci<<std::endl;
pcout<< "time to interation "<<ci<< ": "<<global::timer("simulation").getTime()<<std::endl;
}
if (ci == 30){
writeImages(*lattice,*concentrationLattice,flagMatrix,ci,location,dx,dt,"concentration",flow);
}
// if (ci == pulse_length){
// setBoundaryDensity(*concentrationLattice, inletDomain, (T)0.);
// }
}
}
// ###########################################################################################################
// this the end of hte main run marker
// Running of main code
int main(int argc,char *argv[])
{
plbInit(&argc, &argv);
// for debugging
// For better debugging.
enableCoreDumps();
unbufferOutputStdStreams();
// The try-catch blocks catch exceptions in case an error occurs,
// and terminate the program properly with a nice error message.
// 1. Read command-line parameter: the input file name.
std::string xmlFileName;
try {
global::argv(1).read(xmlFileName);
}
catch (PlbIOException& exception) {
pcout << "Wrong parameters; the syntax is: "
<< (std::string)global::argv(0) << " input-file.xml" << std::endl;
return -1;
}
// 2. Read input parameters from the XML file.
try {
XMLreader document(xmlFileName);
readParameters(xmlFileName);
}
catch (PlbIOException& exception) {
pcout << "Error Reading XML" << exception.what() << std::endl;
return -1;
}
if (onEuler){global::directories().setOutputDir("/cluster/scratch/inaets/palabos/continuefiles/out"+std::to_string(int(flowPercent))+"/");
}else{
global::directories().setOutputDir("./VTK/");
}
// Some clusters have fast parallel input/output facilities
// which Palabos can exploit.
global::IOpolicy().activateParallelIO(useParallelIO);
std::string continueFileName = "";
try {
global::argv(2).read(continueFileName);
}
catch (PlbIOException& exception) { }
// 3. Execute the main program.
try {
run(continueFileName);
}
catch (PlbIOException& exception) {
pcout << exception.what() << std::endl;
return -1;
}
}